Dr. Pierre Zalloua, is a Professor of Genetics and Chair of the Department of Public Health and Epidemiology at Khalifa University. He is also an Adjunct Professor at the Harvard T.H. Chan School of Public Health. He is a population geneticist studying the genetics and other determinants of Diabetes and Cardiovascular Disease. In his research, he has been applying genomic technologies and computational biology, along with detailed clinical and demographic information, to identify Coronary Artery Disease (CAD) and Diabetes Mellitus susceptibility genes. He has established one of the largest CAD databases in the Middle East with more than 10,000 patients with detailed clinical, demographic and biological samples. His database was incorporated into one of the world’s largest CAD consortia (CARDIoGRAM+C4D). He has elucidated the roles of family history and consanguinity in promoting CAD risk and age at diagnosis and demonstrated the compounded impact of known CAD risk factors, such as diabetes, hyperlipidemia, hypertension, smoking, and BMI, on CAD risk and age at diagnosis. Him and his colleagues identified several susceptibility alleles for T2D and CAD in Near Eastern populations. He authored and co-authored more than 170 scholarly publications in his field and has been the recipient of multiple competitive research grants from the European Commission (FP6), QNRF and the NIH. Some of his work has been featured in the New York Times, National Geographic and most international media outlets.
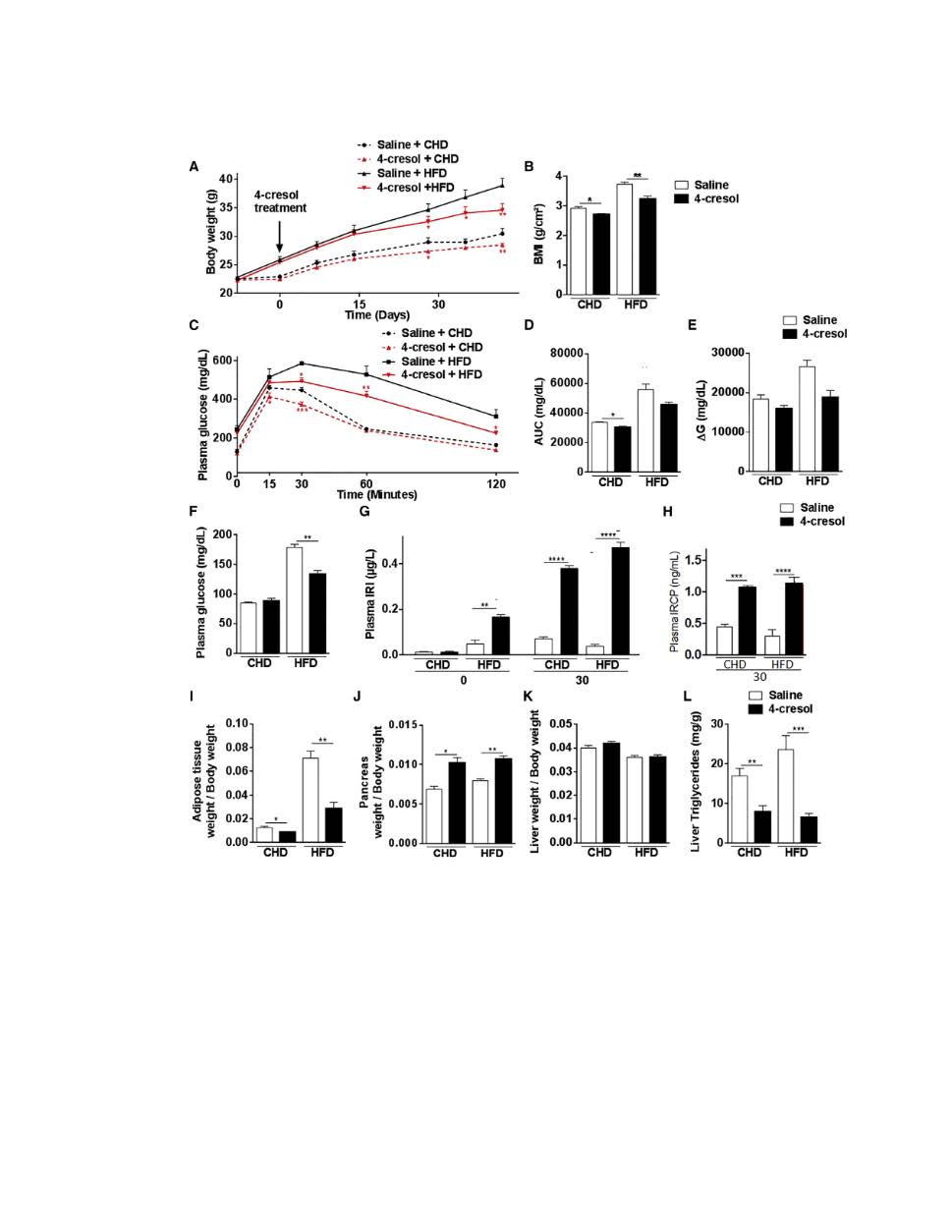
Research and Innovation Grant (RIG-2023:): 2023-2026: Analyzing patterns of single cell gene expression post 4-Cresol treatment.
We have shown that 4-cresol stimulates insulin secretion in in vivo mouse models and in human cells and increases the proliferation of the insulin-producing β-cells. The 4-cresol metabolite likely exerts its function by downregulating the dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A (DYRK1A) and by upregulating the sirtuin 1 gene (SIRT1). These results demonstrate that the microbial metabolite is involved, at least in part, in the inhibition of DYRK1A; We plan to investigate the genes that differentially modulate the expression of DYRK1A and SIRT 1 in the presence or absence of 4-cresol.We will analyze single-cell gene expression pattern changes with or without the presence of non-toxic doses of 4-cresol using Beta islet cells. We will investigate selected genes that mediate interaction between 4-cresol, DYRK1A and SIRT1 and modulate their expression and investigate their impact on glucose homeostasis and insulin secretion in vitro and in preclinical models.
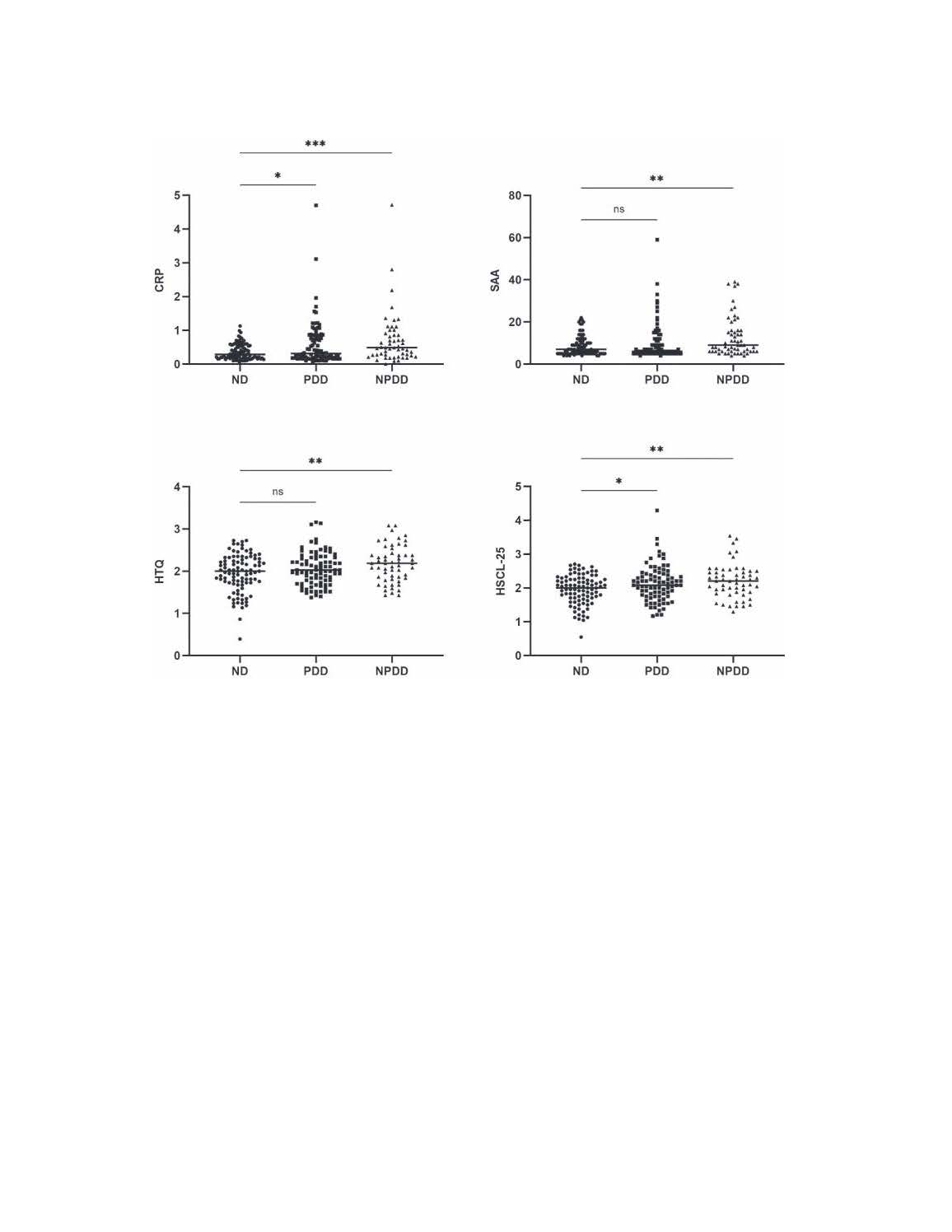
Dissecting the impact of forced displacement on Type 2 Diabetes
The proposed project presents a unique opportunity to create a sustainable research platform through building a unique cohort of individuals with a diverse psychosocial and medical background with a capacity to breed a multitude of interdisciplinary studies investigating the short and long-term health effects of forced displacement on T2D and other chronic NCDs. The proposed project provides an ideal setting to apply analytic tools grounded in a life stage, holistic approach and multidisciplinary strategy that integrates epidemiology and population health sciences to advance our understanding of the determinants of T2D and other NCDs.
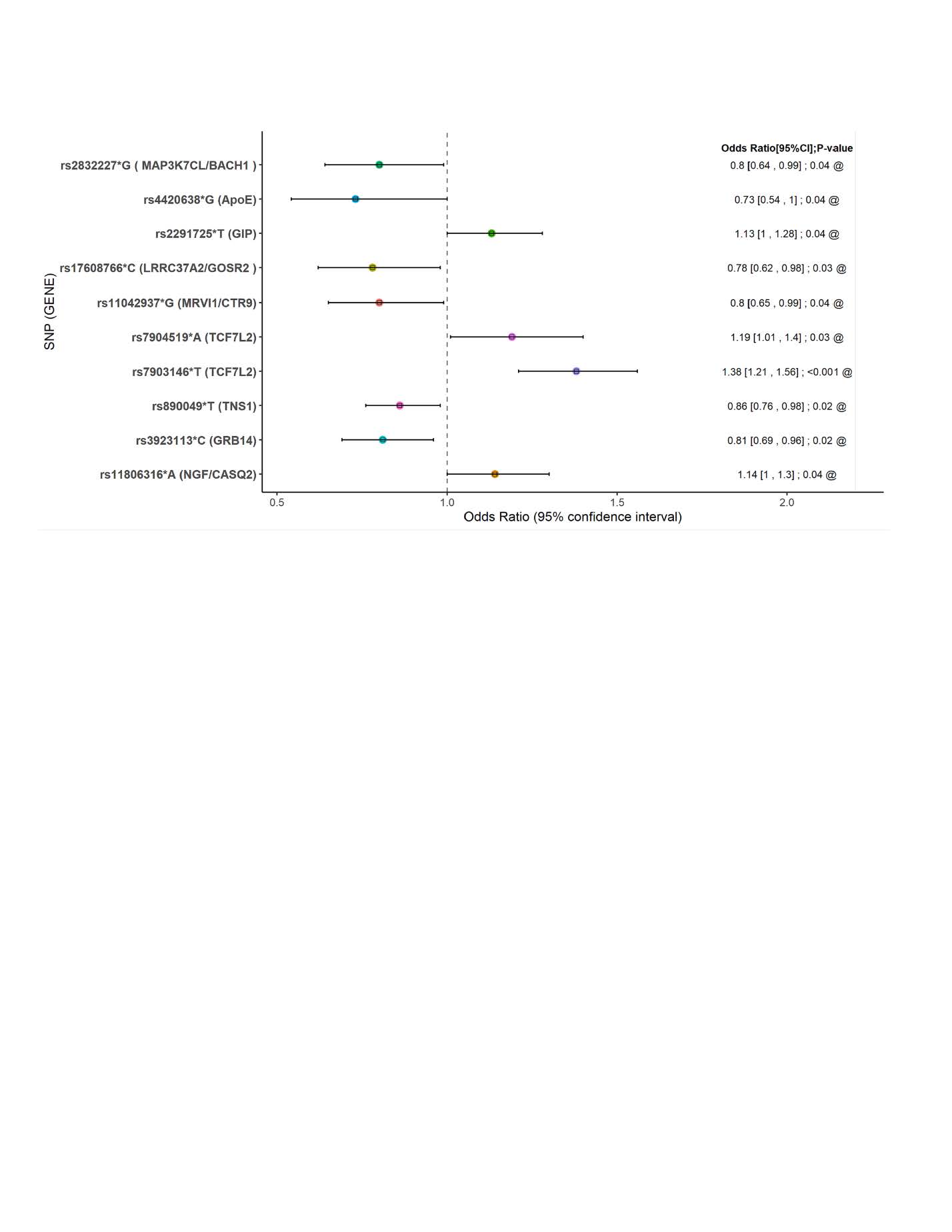
Determinants of coronary artery disease in older diabetic patients.
Considering the large number of T2D and CAD pathogenic pathways, and their limited ability to predict disease causation in any particular individual, common genetic variants and other determinants (e.g., behavioral, or metabolic) will invariably show weak association with disease in a given population. We propose to identify significantly dependent combinatorial phenotypic and genetic associations using modified pattern discovery. In the first instance, we will apply logistic regression to identify signatures or variation patterns predicting phenotype and use these signatures to associate specific genetic variants within targeted inflammatory pathways using currently available data