The features of a cell nucleus can reveal much about the health of a cell, but finding the nucleus among the background noise of a tissue sample can be laborious and time-consuming when done manually. Khalifa University researchers are exploring how AI can be leveraged to speed up the process of detecting a cell’s nucleus.
Known as the cell’s ‘command center’, the nucleus is a large organelle that stores the cell’s DNA. The nucleus controls all of the cell’s activities, such as growth and metabolism, using the DNA’s genetic information.
Pathologists use features of the cell nucleus to distinguish benign from malignant cells. They examine tissue samples for cells with increased nuclear size and irregularities in the nuclear membrane or abnormal distribution of chromatin, a substance within the chromosomes in the nucleus. Being able to automate nuclei detection could serve as a useful tool to make better decisions in cancer diagnosis, prognosis, and treatment.
Artificial intelligence techniques are being investigated to streamline the process of nuclei detection. However, these techniques struggle when the tissue samples are noisy or when nuclei appear crowded.
Dr. Naoufel Werghi, Associate Professor, has collaborated with Dr. Sajid Javed, Assistant Professor, and Dr. Jorge Dias, Professor, to develop a machine learning solution that can sift through the background noise and more easily identify individual nuclei. Their results were published in Medical Image Analysis.
In an ideal world, a sample of tissue would contain only that tissue, but most samples contain a rich mix of several other types. Most normal epithelial cells have nuclei which are round to oval-shaped, most lymphoid cells have round nuclei, while stromal cells have ovoid to spindle-shaped nuclei. Any machine learning algorithm would need to be trained on the specific nucleus shape for the nucleus it is tasked to detect, adding a further layer of training required if the algorithm were to be used in other applications.
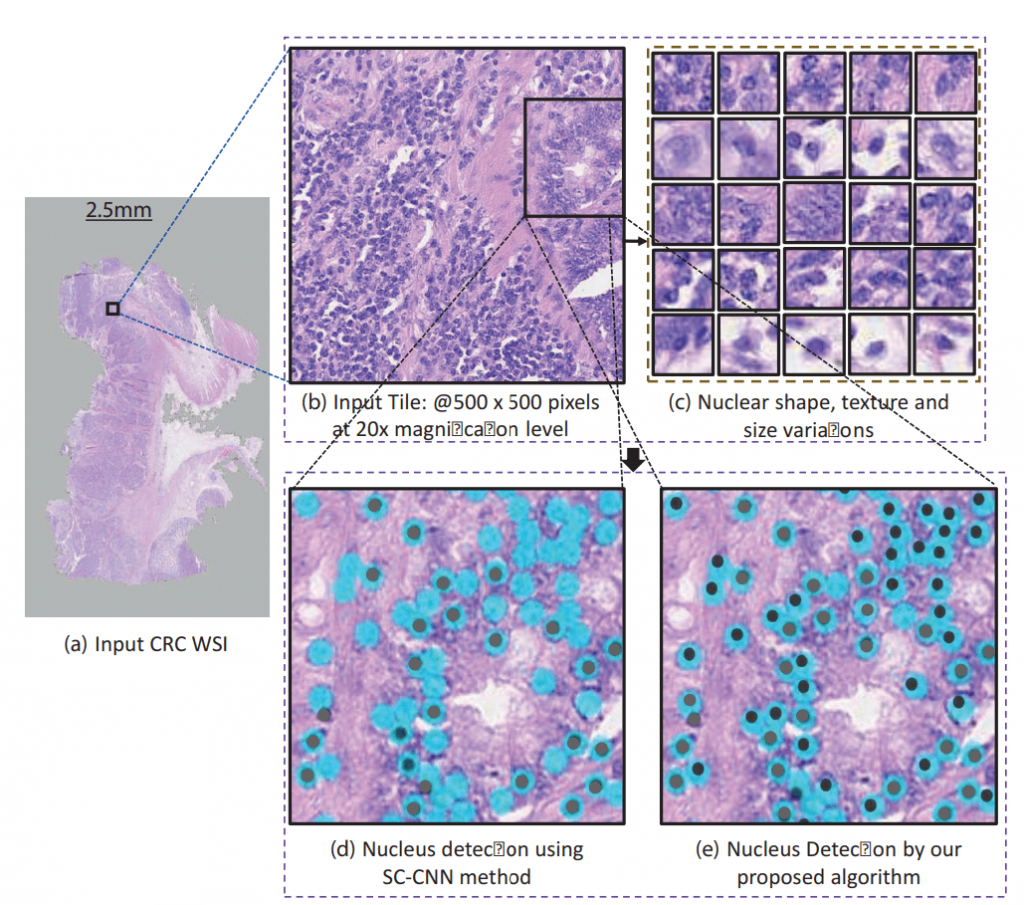
“Nucleus detection is a challenging task because of the nuclear clutter and diverse shapes and sizes,” said Dr. Werghi. “Additionally, computation challenges arise because the images analyzed are multi-gigapixel images, and could contain billions of pixels and tens of thousands of cell nuclei.”
A number of potential methods have been reported for automatic detection of cell nuclei, including deep learning methods to train a convolutional neural network (CNN) to generate probability maps of where the cell nuclei are present.
Existing approaches are promising, but they require a significant amount of training data and expensive platforms to cater to the high computational requirements. The model proposed by the research team can be trained using much smaller training datasets that can be executed on a typical desktop computer.
“Our solution uses correlation filters to sort through the data and identify nuclei,” explained Dr. Werghi. “Compared to end-to-end deep learning in previous methods, correlation filters are computationally effective and require significantly less training data. The correlation filters are also flexible and can detect complex and irregular-shaped nuclei without requiring handcrafted features.”
These correlation filters help the algorithm to better discriminate different nuclear components from the non-nuclear regions and also to discern each nucleus from the remaining nuclei where they are clustered.
Constraints are placed on the spatial structure of the nucleus and its local contextual information in the correlation filter framework to handle varying nuclei shapes, texture, and clutter. The first considers the spatial structure of the nucleus, while the second discriminates between the nucleus and the non-nucleus region. Both of these help to reduce the noise in any given sample.
The team plans to explore the strength of correlation filters in analysis classifying cell nuclei and tissue phenotyping problems, such as cancer detection.
This research was funded by Khalifa University Center for Autonomous Robotic Systems (KUCARS).
Jade Sterling
Science Writer
5 July 2021